You are here
Researchers Reveal the Inner Workings of a Viral DNA-Packaging Motor
A trilogy of papers provides insight into a critical step in how some viruses reproduce
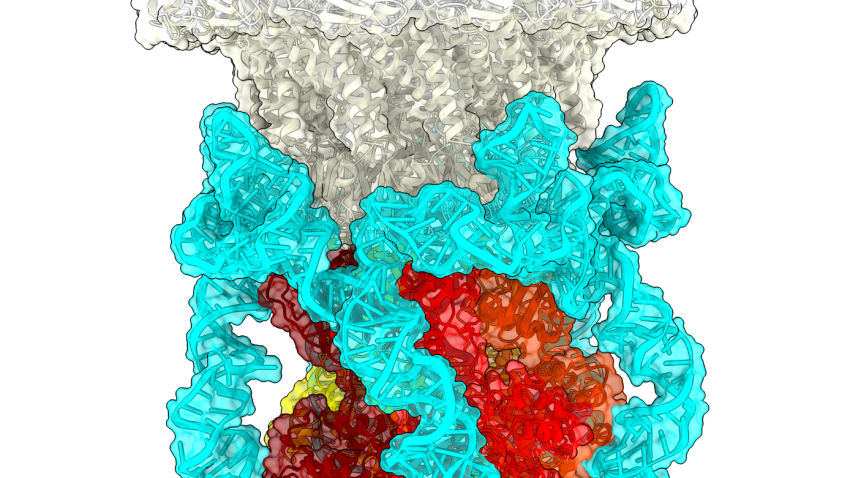
A group of researchers have discovered the detailed inner workings of the molecular motor that packages genetic material into double-stranded DNA viruses. The advance provides insight into a critical step in the reproduction cycle of viruses such as pox- herpes- and adeno-viruses. It could also give inspiration to researchers creating microscopic machines based on naturally occurring biomotors.
The research was conducted by scientists from Duke University, the University of Minnesota, the University of Massachusetts and the University of Texas Medical Branch (UTMB). The results appeared online in a trilogy of papers published in Science Advances, Proceedings of the National Academy of Sciences and Nucleic Acids Research.
“There were several missing pieces of information that prevented us from understanding how these kinds of DNA packaging motors work, which hindered our ability to design therapeutics or evolve new technologies,” said Gaurav Arya, professor of mechanical engineering and materials science, biomedical engineering, and chemistry at Duke. “But with new insights and simulations, we were able to piece together a model of this fantastic mechanism, which is the most detailed ever created for this kind of system.”
Viruses come in many varieties, but their classification generally depends upon whether they encode their genetic blueprints into RNA or single- or double-stranded DNA. The difference matters in many ways and affects how the genetic material is packaged into new viruses. While some viruses build a protein container called a capsid around newly produced RNA or DNA, others create the capsid first and then fill it with the genetic material.
"With new insights and simulations, we were able to piece together a model of this fantastic mechanism, which is the most detailed ever created for this kind of system. Hopefully it will inspire other researchers to create new inventions using these same mechanisms.”
Gaurav Arya, professor of mechanical engineering & Materials science
Most double-stranded DNA viruses take the latter route, which presents many challenges. DNA is negatively charged and does not want to be crammed together into a small space. And it’s packaged into an extremely dense, nearly crystalline structure, which also requires a lot of force.
“The benefit of this is that, when the virus is ready to infect a new cell, the pressure helps inject DNA into the cell once it’s punctured,” said Joshua Pajak, a doctoral student working in Arya’s laboratory. “It’s been estimated that the pressure exceeds 800 PSI, which is almost 10 times the pressure in a corked bottle of champagne.”
Forcing DNA into a tiny capsid at that amount of pressure requires an extremely powerful motor. Until recently, researchers only had a vague sense of how that motor worked because of how difficult it is to visualize. The motor only assembles on the virus particle, which is enormous compared to the motor.
“Trying to see the motor attached to the virus is like trying to see the details in the Statue of Liberty’s torch by taking a photo of the entire statue,” said Pajak.
But at a recent conference, Pajak learned that Marc Morais, professor of biochemistry and molecular biology at UTMB, and Paul Jardine, professor of diagnostic and biological sciences at the University of Minnesota, had been working on this motor for years and had the equipment and skills needed to see the details. Some of their initial results appeared to match the models Pajak was building with what little information was already available. The group grew excited that their separate findings were converging toward a common mechanism and quickly set about solving the mystery of the viral motor together.
In a paper published in Science Advances, Morais and his colleagues resolved the details of the entire motor in one of its configurations. They found that the motor is made up of five proteins attached to one another in a ring-like formation. Each of these proteins are like two suction cups with a spring in between, which allows the bottom portion to move vertically in a helical formation so that it can grab onto the helical backbone of DNA.
“Because you could fit about 100,000 of these motors on the head of a pin and they’re all jiggling around, getting a good look at them proved difficult,” said Morais. “But after my UTMB colleagues Michael Woodson and Mark White helps us image them with a cryo-electron microscope, a general framework of the mechanism fell into place.”
 In a second paper, published in Nucleic Acids Research, the Morais group captured the motor in a second configuration using x-ray crystallography. This time the bottom suction cups of the motor were all scrunched up together in a planar ring, leading the researchers to imagine that the motor might move DNA into the virus by ratcheting between the two configurations.
In a second paper, published in Nucleic Acids Research, the Morais group captured the motor in a second configuration using x-ray crystallography. This time the bottom suction cups of the motor were all scrunched up together in a planar ring, leading the researchers to imagine that the motor might move DNA into the virus by ratcheting between the two configurations.
To test this hypothesis, Pajak and Arya performed heavy-duty simulations on Anton 2, the fastest supercomputer currently available for running molecular dynamics simulations. Their results not only supported the proposed mechanism, but also provided information on how exactly the motor’s cogs contort between the two configurations.
While the tops of the proteins remain statically attached to the virus particle, their bottom halves move up and down in a cyclic pattern powered by an energy-carrying molecule called ATP. Once all the proteins have moved up—dragging the DNA along with them—the proteins release the byproduct of the ATP chemical reaction. This causes the lower ring to release the DNA and reach back down into their original helical state, where they once again grab on to more ATP and DNA to repeat the process.
“Joshua pieced together lots of clues and information to create this model,” said Arya. “But a model is only useful if it can predict new insights that we didn’t already know.”
At its core, the model is a series of mechanical actions that must fit together and take place in sequential order for everything to work properly. Pajak’s simulations predicted a specific series of mechanical signals that tell the bottoms of the proteins whether or not they should be gripping the DNA. Like a line of dominoes falling, removing one of the signaling pathways from the middle should stop the chain reaction and block the signal.
To validate this prediction, the researchers turned to Jardine and colleagues Shelley Grimes and Dwight Anderson to see if removing one of the signaling dominoes actually stopped the motor from packaging DNA. A third paper, published in PNAS, shows that the sabotage worked. After mutating a domino in the signaling pathway so that it was unable to function, the motor could still bind and burn fuel just as well as ever, but it was much worse at actually packaging DNA.
“The new mechanism predicted by the high-resolution structures and the detailed predictions provided a level of detail greater than we ever previously had,” said Jardine. “This allowed us to test the role of critical components of the motor, and therefore assess the validity of this new mechanism as we currently understand it.”
The result is a strong indication that the model is very close to describing how the motor behaves in nature. The group plans to continue their highly integrated structural, biochemical and simulation approach to further test and refine the proposed model. They hope that that this fundamental understanding could potentially be used to someday fight disease or create a synthetic molecular motor.
“All technology is inspired by nature in one way or another,” said Arya. “Now that we really know how this molecular motor works, hopefully it will inspire other researchers to create new inventions using these same mechanisms.”
This work was supported by the National Institutes of Health (GM118817, GM122979, GM127365) and the National Science Foundation (MCB1817338). Cryo-EM data were also collected at NIH-supported regional cryo-EM imaging facilities (1U24 GM116787-01, 1U24 GM116792-01). Simulations were performed on Anton 2 supercomputer made available by D.E. Shaw Research and hosted at the Pittsburgh Supercomputing Center via the NIH (R01GM116961) and the Comet supercomputer hosted at the San Diego Supercomputer Center via the NSF (ACI-1053575).
CITATIONS:
“Viral Packaging ATPases Utilize a Glutamate Switch to Couple ATPase Activity and DNA Translocation.” Joshua Pajak, Rockney Atz, Brendan J. Hilbert, Marc C. Morais, Brian A. Kelch, Paul Jardine, and Gaurav Arya. PNAS, April 27, 2021 118 (17) e2024928118. DOI: 10.1073/pnas.2024928118
“A Viral Genome Packaging Motor Transitions Between Cyclic and Helical Symmetry to Translocate dsDNA,” Michael Woodson, Joshua Pajak, Bryon P. Mahler, Wei Zhao, Wei Zhang, Gaurav Arya, Mark A. White, Paul J. Jardine and Marc C. Morais. Science Advances, 07 May 2021: Vol. 7, no. 19, eabc1955. DOI: 10.1126/sciadv.abc1955
“Atomistic Basis Of Force Generation, Translocation, and Coordination in a Viral Genome Packaging Motor,” Joshua Pajak, Erik Dill2, Emilio Reyes-Aldrete, Mark A. White, Brian A.Kelch, Paul J. Jardine, Gaurav Arya1, and Marc C. Morais. Nucleic Acids Research, 2021. DOI: 10.1093/nar/gkab372
